Abstract
Breast cancer stem cells: From beginning
Cancer stem cells (CSCs) in breast tumors were first discovered by Al-Hajj et al. from the University of Michigan in 2003 Al-Hajj, et al., 2003 . They cultured primary breast tumors and analyzed the tumor cells for the expression of CD24 + , CD24, and ESA. The results showed that only cancer cells with CD44 + CD24 - /lowESA+ phenotype were highly tumorigenic. In contrast, cells with CD24 + -CD24+phenotype did not induce tumor formation. Moreover, they also observed that CD44 + CD24 - /lowESA+ cells could maintain their tumorigenic potential when serially passaged for long-term. Based on these results, Al-Hajj et al. concluded that CD44 + CD24 - /lowcells satisfied the criteria for CSCs, such as self-renewal, differentiation, and high tumorigenicity. These results were further confirmed by several other studies Cho, et al., 2008 Liang, et al., 2013 Pece, et al., 2010 Pham, et al., 2011 Ponti, et al., 2005 Saadin and White, 2013 Vassilopoulos, et al., 2008 Walia and Elble, 2010 .
Where are breast CSCs from?
Breast CSCs are formed through these mechanisms. Some in vitro studies and clinical investigations suggest that breast CSCs are produced in vivo through the following 2 mechanisms: (1) accumulation of mutations in mammary stem cells during development that causes these cells to lose their self-renewal capacity and (2) development of breast CSCs from non-stem cells through different evolutionary mechanisms.
Increased risk of breast cancer in children exposed to radiationindicates that breast cancer may develop directly from long-lived stem or progenitor cells present in the mammary glands Miller, et al., 1989 Modan, et al., 1989 . Importantly, luminal progenitor cells are known to induce the formation of BRCA1-driven tumors Lim, et al., 2009 Molyneux, et al., 2010 Proia, et al., 2011 . Pece et al. (2010) showed that poorly differentiated cancers had higher number of CSCs than well-differentiated cancers Pece, Tosoni, Confalonieri, Mazzarol, Vecchi, Ronzoni, Bernard, Viale, Pelicci and Di Fiore, 2010 ( Figure 1 ).
Figure 1.
In a study involving 61 patients with breast cancer, normal breast tissue and breast tumor tissue from the same patients were analyzed to determine the presence of stem cells in both the tissues. In all, nine patients had triple-negative cancer (TNC) and 52 patients had ER+and Her2+cancer. The results showed that 100% (9/9) patients with TNC had CSCs with CD44 + CD49f+CD133/2+phenotypein both the normal and tumor tissues and that only 13.4% (7/52) patients with ER+ and/or Her2+cancerhad CSCs in the normal breast tissue. Based on this result, Atkinson et al. (2013) proposed that CSCs in breast cancer originated from mutated mammary stem cells present in the normal breast tissue Atkinson, et al., 2013 .
The idea that breast CSCs were derived from non-stem cells originated from the heterogeneity of breast cancer cell lines. Almost all breast cancer cell lines contain a subpopulation of cells with breast CSC phenotypes. Interestingly, some studies have shown that different subpopulations of cells in the same breast cancer cell line can interconvert between phenotypes.
Analysis of some commercial breast cancer cell lines such as SUM159, SUM149, Ca1a, MDA-MB-231,BT474,SKBR3,andMCF7 indicated that these cell lines included a small population of stem cells that underwent self-renewal to form mammospheres in serum-free cultures and differentiation to generate other cells that resulted in heterogeneity Gupta, et al., 2011 Meyer, et al., 2009 Piggott, et al., 2011 .
Interconversion of CSCs in breast cancer cell lines was clearly recorded by Gupta et al. (2011). They sorted breast CSCs with stem-like basal and luminal phenotypes from 2 breast cancer cell lines SUM159 and SUM149and observed that these cells exhibited the properties of their parental cell lines after 11 days in culture Gupta, Fillmore, Jiang, Shapira, Tao, Kuperwasser and Lander, 2011 . In another study, Piggott et al. (2011) depleted CSCs from BT474 breast cancer cell line and observed that this depleted cell line re-established progenitor-like cells after4weeksinculture Piggott, Omidvar, Marti Perez, Eberl and Clarkson, 2011 . In some cell lines such asCa1a, MCF7, SUM159, and MDA-MB-231, breast cells without a CSC phenotype (CD44 + CD24+) produced breast CSCs with CD44 + CD24 - phenotype in vitro Meyer, Fleming, Ali, Pesesky, Ginsburg and Vonderhaar, 2009 . In addition, non-CSCs (CD44 + CD24+ phenotype) isolated from Ca1a,ZR75-1,and MCF7cell lines successfully produced heterogeneous tumors showing local invasion in immuno compromisedmice Meyer, Fleming, Ali, Pesesky, Ginsburg and Vonderhaar, 2009 . These evidences confirmed that breast non-CSCs could be automatically converted into CSCs both in vitro and in vivo ( Figure 1 ).
Some recent studies have shown that actively transdifferentiated breast non-CSCs can be converted into CSCs after treatment with some defined factors. CSC-like cells were produced after the transfection of MCF10Acells with SRC oncogene Iliopoulos, et al., 2011 . Moreover, the CSC-conditioned medium can convert non-CSCs into CSCs Iliopoulos, Hirsch, Wang and Struhl, 2011 . Shaffer et al. also produced CSCs by transfecting non-CSCs withSV40andH-ras Chaffer, et al., 2011 .
Some studies have shown that breast CSCs can be produced in vitro by fusing breast cancer cells with mesenchymal stromal cells or macrophages. Hybrids of bone marrow-derived mesenchymal stem cells (MSCs) and cells from 2 breast cancer cell lines MDA-MB-231 (MDA) and MA11 show increased metastatic capacity. Because MSCs can migrate and localize to breast cancers, formation of MSC–breast cancer cell hybrids maybe a potential mechanism for generating breast CSCs Rappa, et al., 2012 . Fusion of M2 macrophages with cells from breast cancer cell lines MCF-7 and MDA-MB-231 in the presence of polyethylene glycol produces hybrids that show increased migration, invasion, and tumorigenicity and haveCD44 + CD24 - /low phenotype Ding, et al., 2012 .
What the markers of breast cancer stem cells?
Specific markers
Studies have shown that BCSCs are presentin tumors and are defined by a unique set of markers. Many previously published reports have described the isolation of BCSCs from malignant tumors using specific markers ( Ponti, Costa et al. 2005 ; Pham, Phan et al. 2011 ; Lin, Hutzen et al. 2013; Wang, Lv et al. 2014). However, other studies have shown that BCSCs exists a heterogeneous population in tumors (Lorico and Rappa 201; Hwang-Verslues, Lee et al. 2012; Wong, Fuller et al. 2012). This indicated that BCSCs could express different markers with different levels of expression at various times. This was consistent with other theories about cancer stem cells (CSCs).
There are two theories about stem cell heterogeneity. The first is related to the existence of different CSCs in different tumors. The second is related to the theory of clonal evolution, which describes how different tumors can originate from a single stem cell. A recent report showed that maybe there is a dynamic balance between cancer progenitor cells and CSCs (Li and Laterra 2012). A CSC can differentiate into a progenitor cancer cell, and a progenitor cancer cell can de-differentiate into a CSC. This suggested that CSCs and their progenitor cancer cells co-exist in a dynamic form (Li and Laterra 2012). The relationship between CSCs and cancer progenitor cells creates the necessity to specify a combination of markers for the identification of CSCs.
Heterogeneity in the BCSC population: which is the strongest sub-population?
Several recent studies have shown that BCSCs contain sub-populations that exhibit different tumorigenicity. To date, only some of these subpopulations have been identified. Ghebeh et al. determined that there are 3 main cell populations in the normal breast tissue, including, “basal A” progenitor cells with the markers Ep-CAM-/lowandCD49f+, “luminal B” progenitor cells with the markers Ep-CAMhighandCD49f+, and “luminal C” differentiated cells with the markers Ep-CAMhighandCD49f- (Ghebeh, Sleiman et al. 2013). These populations have been culturedin order to identify their mammosphere forming potential. The results show that only subpopulations basal A and luminal B could form a mammosphere whereas luminal C could not form a mammosphere. Moreover, the mammosphere-forming capacity of basal A is stronger than that of luminal B. It was shown that only subpopulations of basal A and luminal B contain small populations of cells with CD24 + high and CD24 low . This suggested that in the normal breast tissue there are at least two subpopulations of cells with markers CD24 + high andCD24 low .
Similarly, there are three populations of cells in breast cancer tissue, including, basal A, luminal B, and luminal C. However, there are differences in their ratios. In fact, the number of basal A cells significantly decreases as the number of luminal C cells increases. In contrast to normal tissue, in cancer tissue both luminal B and luminal C cells contain the markers CD24 + high andCD24 low . However, CD24 + high andCD24 low cells of luminal B can form mammospheres more easily than luminal C cells. In comparison to subpopulations, MUC-1-, ALDH + , and CD10+, CD24 + high CD24 low cells of luminal B have the greatest mammosphere-forming capacity. In summary, the CD24 + high CD24 low cell population from a breast tumor can contain subpopulations that differ in the expression of Ep-CAM and CD49f. Hence, the results of this analysis indicate that CD24 + high CD24 low Ep-CAM+CD49f+could be a marker profile for BCSCs.
Molyneux et al. (2010) showed that breast cancer cells originate from luminal epithelial progenitors and not from basal stem cells Molyneux, Geyer et al. 2010 . Uchoa Dde et al. (2014) determined that breast cancer cells express the BCSC phenotype (Uchoa Dde, Graudenz et al. 2014). In a previous study, immunohistochemistry was used to show that there were BCSCs in hormone-receptor-positive breast cancer, but there was no correlation between markers of CSCs and the response to endocrine therapy and overall clinical outcome (Hashimoto, Shimizu et al. 2012).
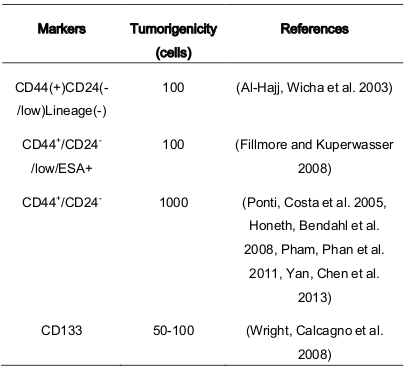
The heterogeneity of the BCSC population based on cell markers CD24 + high CD24 low has resulted in conflicting conclusions. For example, Lin et al. (2012) showed that there is a correlation between of prevalence of CD44 + CD24 - tumor cells and invasive ductal carcinoma, high recurrence, and shorter DFS and OS. Therefore, this marker is important for breast cancer diagnosis and treatment. Conversely, Zhong et al. (2014) suggested that ALDH + is a better clinical indicator for relapse and invasive ductal carcinoma than CD44 + CD24 - ( Table 1 ).
ALDH
Aldehyde dehydrogenases (ALDHs) are a group of enzymes that catalyze the oxidation (dehydrogenation) of aldehydes (Marchitti, Brocker et al. 2008). There are 19 kinds of ALDH identified in humans. One of these is ALDH1A, a well-known enzyme responsible for oxidizing aldehydes to carboxylic acids (Marchitti, Brocker et al. 2008). ALDH1A is highly expressed in the epithelium of testis, brain, eye, liver, kidney, and some stem cells. To date, ALDH1A exhibits two main functions in hematopoietic stem cells and neural stem cells. The first is the oxidation of retinal to retinoic acid (Collins 2008). The retinoic acid activates nuclear retinoic acid receptors (RARs) which in turn regulate the expression of related genes. Second, ALDH1A can metabolize and detoxify chemotherapeutics (Magni, Shammah et al. 1996). Due to this property, ALDH1A is known as a contributing factor to chemotherapy resistance of hematopoietic stem cells.
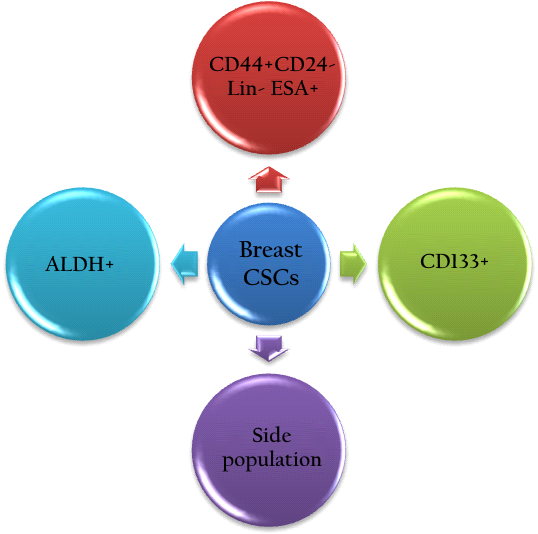
In the recent years, ALDH1A has been thought to be expressed in a sub-population of breast cancer cells ( Figure 2 ). These ALDH + breast cancer cells exhibit stem cell properties (Ginestier, Hur et al. 2007). Clinically, ALDH + is suggested to be a prognostic marker to predict metastasis and poor patient outcome (Balicki 2007; Ginestier, Hur et al. 2007; Charafe-Jauffret, Ginestier et al. 2010; Marcato, Dean et al. 2011; Liu, Lv et al. 2014). In fact, ALDH1A expression in breast tumors is associated with chemotherapy resistance (Tanei, Morimoto et al. 2009; Alamgeer, Ganju et al. 2014).
What is the difference between the three populations considered to be BCSCs? There is a population with the phenotype of CD44 + CD24 - , another with ALDH, and another with CD133. There are also combinations of phenotypes such as CD44 + CD24 - , CD133 + , and ALDH + . Some populations of BCSCs also include phenotypes CD44 + CD133 + , CD44 + ALDH + , or CD133 + ALDH + . These differences seem to be related to distinct levels of differentiation status in BCSCs (Ricardo, Vieira et al. 2011).
Zhong et al. (2014) demonstrated that ALDH1 + and CD44 + /CD24 - breast cancer cells play significant roles in metastasis. However, the ALDH1 marker is a better predictive marker for breast cancer metastasis than the CD44 + /CD24 - phenotype (Zhong, Shen et al. 2014). In a previous study, Tanei et al. (2009) found that ALDH1-positive but not CD44 + /CD24 - BCSCs played a significant role in resistance to chemotherapy (Tanei, Morimoto et al. 2009). This marker was also highly expressed in TNBC compared to non-TNBC Li, Ma et al. 2013 .
Side population
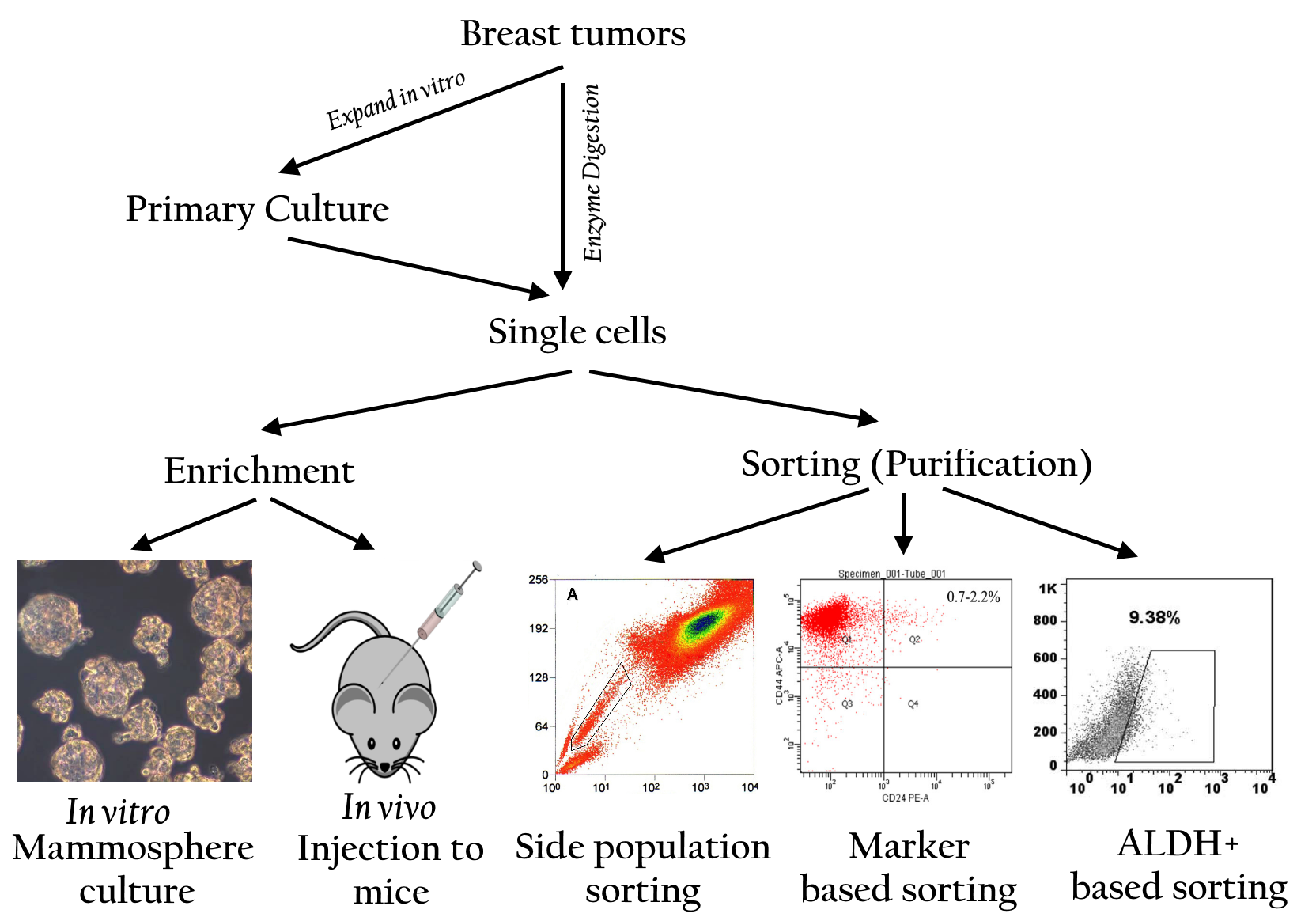
A side population (SP) is defined as a subpopulation of cells that expresses distinct properties compared to the main population in flow cytometry analysis. The SP cell phenotype was first discovered in 1996 in mouse bone marrow. SP cells in bone marrow are related to hematopoietic stem cells (Goodell, Brose et al. 1996). Using this established technique, some authors have also detected SP cells in other normal tissues including the mammary gland (Alvi, Clayton et al. 2003) and tumors including breast tumor (Britton, Kirby et al. 2011). In the stem cell analysis, a side population is defined as a minor cell population that exhibits low-intensity staining with dyes such as Hoechst 33342 (H33342) or rhodamine 123 (R123). Both H33342 and R123 easily stain cells because they can cross the cell membrane. However, SP cells absorb less of the dye. Some investigations have suggested that side population cells over-expressed trans-membrane transporters like ATP-binding cassette (ABC) molecule ABCG2/BCRP (ATP-binding cassette, sub-family G/breast cancer resistance protein-1). Due to the high expression of this ABC transporter, SP cells can extrude some dyes and drugs (Hadnagy, Gaboury et al. 2006; Wu and Alman 2008). Therefore, this population is clearly visible in flow cytometry analysis. SPs are present in both normal breast tissue and breast cancer tissue but are limited in number. These cells are characterized as having some of the same properties as stem cells. However, it has been shown that SPs exist in almost all breast cancer cell lines (Patrawala, Calhoun et al. 2005).
What is the relationship between SP cells and the cancer stem cell phenotype? To date, whether the SP cell population is enrichedby cancer stem cells remains a controversial topic. Some studies have showed that SPs contain cells with higher clonogenic and tumorigenic potential than non-SPs (Patrawala, Calhoun et al. 2005; Hiraga, Ito et al. 2011). However, other studies have claimed that SP cells do not relate to cancer stem cells and, more importantly, that the cancer stem cell population is outside of the SP cells. It is thought that cancer stem cells highly express ABCG2, which contributes to anti-tumor drug resistance and that cancer stem cells are SP cells. However, Patrawala et al. (2005) demonstrated that ABCG2 cancer cells also exhibit high clonogenic and tumorigenic potential, while also expressing several genes for “stemness” (Patrawala, Calhoun et al. 2005) ( Figure 3 ).
References
- M Al-Hajj, MS Wicha, A Benito-Hernandez, SJ Morrison, MF Clarke. Prospective identification of tumorigenic breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3983-3988. Google Scholar
- RL Atkinson, WT Yang, DG Rosen, MD Landis, H Wong, MT Lewis, CJ Creighton, KR Sexton, SG Hilsenbeck, AA Sahin, AM Brewster, WA Woodward, JC Chang. Cancer stem cell markers are enriched in normal tissue adjacent to triple negative breast cancer and inversely correlated with DNA repair deficiency. Breast cancer research. 2013;BCR 15:R77. Google Scholar
- CL Chaffer, I Brueckmann, C Scheel, AJ Kaestli, PA Wiggins, LO Rodrigues, M Brooks, F Reinhardt, Y Su, K Polyak, LM Arendt, C Kuperwasser, B Bierie, RA Weinberg. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7950-7955. Google Scholar
- RW Cho, X Wang, M Diehn, K Shedden, GY Chen, G Sherlock, A Gurney, J Lewicki, MF Clarke. Isolation and molecular characterization of cancer stem cells in MMTV-Wnt-1 murine breast tumors. Stem cells (Dayton, Ohio). 2008;26:364-371. Google Scholar
- J Ding, W Jin, C Chen, Z Shao, J Wu. Tumor associated macrophage x cancer cell hybrids may acquire cancer stem cell properties in breast cancer. PloS one. 2012;7:e41942. Google Scholar
- PB Gupta, CM Fillmore, G Jiang, SD Shapira, K Tao, C Kuperwasser, ES Lander. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633-644. Google Scholar
- D Iliopoulos, HA Hirsch, G Wang, K Struhl. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1397-1402. Google Scholar
- S Liang, M Furuhashi, R Nakane, S Nakazawa, H Goudarzi, J Hamada, H Iizasa. Isolation and characterization of human breast cancer cells with SOX2 promoter activity. Biochemical and biophysical research communications. 2013;437:205-211. Google Scholar
- E Lim, F Vaillant, D Wu, NC Forrest, B Pal, AH Hart, ML Asselin-Labat, DE Gyorki, T Ward, A Partanen, F Feleppa, LI Huschtscha, HJ Thorne, SB Fox, M Yan, JD French, MA Brown, GK Smyth, JE Visvader, GJ Lindeman. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nature medicine. 2009;15:907-913. Google Scholar
- MJ Meyer, JM Fleming, MA Ali, MW Pesesky, E Ginsburg, BK Vonderhaar. Dynamic regulation of CD24 and the invasive, CD24+posCD24neg phenotype in breast cancer cell lines. Breast cancer research. 2009;BCR 11:R82. Google Scholar
- AB Miller, GR Howe, GJ Sherman, JP Lindsay, MJ Yaffe, PJ Dinner, HA Risch, DL Preston. Mortality from breast cancer after irradiation during fluoroscopic examinations in patients being treated for tuberculosis. The New England journal of medicine. 1989;321:1285-1289. Google Scholar
- B Modan, A Chetrit, E Alfandary, L Katz. Increased risk of breast cancer after low-dose irradiation. Lancet. 1989;1:629-631. Google Scholar
- G Molyneux, FC Geyer, FA Magnay, A McCarthy, H Kendrick, R Natrajan, A Mackay, A Grigoriadis, A Tutt, A Ashworth, JS Reis-Filho, MJ Smalley. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell stem cell. 2010;7:403-417. Google Scholar
- S Pece, D Tosoni, S Confalonieri, G Mazzarol, M Vecchi, S Ronzoni, L Bernard, G Viale, PG Pelicci, Fiore Di. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62-73. Google Scholar
- PV Pham, NL Phan, NT Nguyen, NH Truong, TT Duong, DV Le, KD Truong, NK Phan. Differentiation of breast cancer stem cells by knockdown of CD24+: promising differentiation therapy. Journal of translational medicine. 2011;9:209. Google Scholar
- L Piggott, N Omidvar, Perez Marti, M Eberl, RW Clarkson. Suppression of apoptosis inhibitor c-FLIP selectively eliminates breast cancer stem cell activity in response to the anti-cancer agent, TRAIL. Breast cancer research. 2011;BCR 13:R88. Google Scholar
- D Ponti, A Costa, N Zaffaroni, G Pratesi, G Petrangolini, D Coradini, S Pilotti, MA Pierotti, MG Daidone. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer research. 2005;65:5506-5511. Google Scholar
- TA Proia, PJ Keller, PB Gupta, I Klebba, AD Jones, M Sedic, H Gilmore, N Tung, SP Naber, S Schnitt, ES Lander, C Kuperwasser. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell stem cell. 2011;8:149-163. Google Scholar
- G Rappa, J Mercapide, A Lorico. Spontaneous formation of tumorigenic hybrids between breast cancer and multipotent stromal cells is a source of tumor heterogeneity. The American journal of pathology. 2012;180:2504-2515. Google Scholar
- K Saadin, IM White. Breast cancer stem cell enrichment and isolation by mammosphere culture and its potential diagnostic applications. Expert review of molecular diagnostics. 2013;13:49-60. Google Scholar
- A Vassilopoulos, RH Wang, C Petrovas, D Ambrozak, R Koup, CX Deng. Identification and characterization of cancer initiating cells from BRCA1 related mammary tumors using markers for normal mammary stem cells. International journal of biological sciences. 2008;4:133-142. Google Scholar
- V Walia, RC Elble. Enrichment for breast cancer cells with stem/progenitor properties by differential adhesion. Stem cells and development. 2010;19:1175-1182. Google Scholar
 Biomedpress
Biomedpress Open Access
Open Access 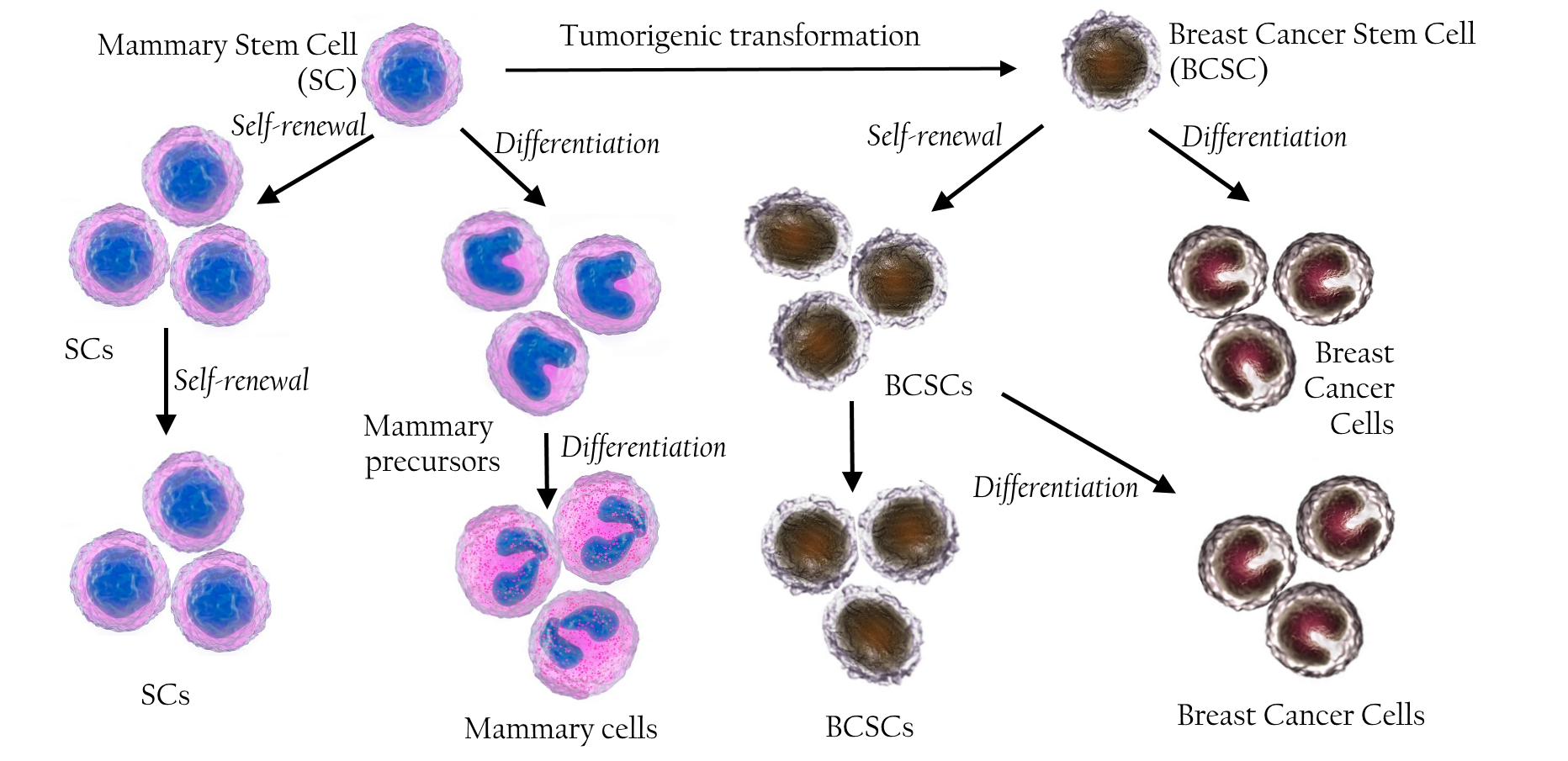







Relations of breast cancer stem cells and normal mammary stem cells